IriaSantos-web
AD3: conexión con la API del COVID-19 y análisis con Pandas
Me conecto a la API https://api.covid19api.com/
Instalación e importación de Pandas
!pip install pandas
Requirement already satisfied: pandas in c:\users\rnasa\anaconda3\lib\site-packages (0.25.1)
Requirement already satisfied: python-dateutil>=2.6.1 in c:\users\rnasa\anaconda3\lib\site-packages (from pandas) (2.8.0)
Requirement already satisfied: numpy>=1.13.3 in c:\users\rnasa\anaconda3\lib\site-packages (from pandas) (1.16.5)
Requirement already satisfied: pytz>=2017.2 in c:\users\rnasa\anaconda3\lib\site-packages (from pandas) (2019.3)
Requirement already satisfied: six>=1.5 in c:\users\rnasa\anaconda3\lib\site-packages (from python-dateutil>=2.6.1->pandas) (1.12.0)
import pandas as pd
Variables/objetos
Variable de nombre url para llamar a la lista de diccionarios de países de la API del Covid-19. Posteriormente invoco la url para saber si ha funcionado la llamada.
url = 'https://api.covid19api.com/countries'
url
'https://api.covid19api.com/countries'
Añado una variable para la función pd.read_json para la variable anterior.
Se podría hacer:
df = pd.read_json(https://api.covid19api.com/countries
Se hace a través de variable para facilitar su uso posterior con código más breve.
Posteriormente invoco el datajson para ver si ha funcionado la importación, lo que muestra una tabla en la que hay 248 elementos. En este caso empieza a contar en 0 por tratarse de json.
Este código nos muestra un estracto con las filas del inicio y las del final.
df = pd.read_json(url)
df
| Country | Slug | ISO2 | |
|---|---|---|---|
| 0 | Gibraltar | gibraltar | GI |
| 1 | Oman | oman | OM |
| 2 | France | france | FR |
| 3 | Jersey | jersey | JE |
| 4 | Mali | mali | ML |
| ... | ... | ... | ... |
| 243 | Puerto Rico | puerto-rico | PR |
| 244 | Papua New Guinea | papua-new-guinea | PG |
| 245 | Saint Pierre and Miquelon | saint-pierre-and-miquelon | PM |
| 246 | Timor-Leste | timor-leste | TL |
| 247 | Montenegro | montenegro | ME |
248 rows × 3 columns
Para visualizar los datos de España, dado que tengo una columna de países, creo una lista nueva que seleccione solo los identificadores similares a Spain.
df[df['Country'] == 'Spain']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 141 | Spain | spain | ES |
Defino otra variable con los datos en tiempo real de España. (¡ATENCIÓN! la nomenclatura con guion bajo, ya que el guion medio no debe emplearse para código de este tipo).
Creo el objeto df_rt_es para que lea la información de la url previa.
Posteriormente muestro los datos para ver que ha funcionado correctamente.
url_rt_es = 'https://api.covid19api.com/country/spain/status/confirmed/live'
df_rt_es = pd.read_json(url_rt_es)
df_rt_es
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 891 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-01 00:00:00+00:00 | |||
| 892 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Spain | ES | 40.46 | -3.75 | 12890002 | confirmed | 2022-07-05 00:00:00+00:00 |
896 rows × 10 columns
Marco df_rt_es.tail() para ver la cola de la tabla.
df_rt_es.tail()
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 891 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-01 00:00:00+00:00 | |||
| 892 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Spain | ES | 40.46 | -3.75 | 12818184 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Spain | ES | 40.46 | -3.75 | 12890002 | confirmed | 2022-07-05 00:00:00+00:00 |
Marco df_rt_es.head() para ver la parte inicial.
df_rt_es.head()
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Spain | ES | 40.46 | -3.75 | 0 | confirmed | 2020-01-26 00:00:00+00:00 |
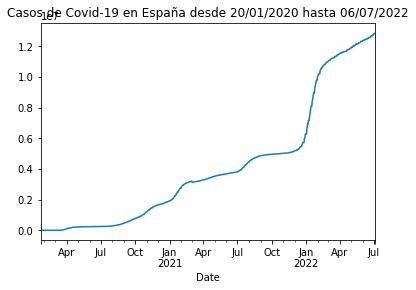
Tenemos una tabla con columnas para ID, país, código de país, provincia, ciudad, código de ciudad, latitud, longitud, número de casos, estado y la fecha. Si quiero obtener una gráfica con el número de casos y la fecha lo que debo hacer es transformar la columna de control. Para ello convierto la columna de fecha en columna de control pidiéndole ya que me muestre los casos España desde el 20 de enero de 2020 a la actualidad con la expresión df_rt_es.set_index('Date')['Cases'].plot(title="Casos de Covid-19 en España desde 20/01/2020 hasta 29/06/2022") a la que debo darle un nombre de variable si no quiero tener problemas con la configuración de Anaconda3. Le ponto de nombre plot_rt_es.
plot_rt_es = df_rt_es.set_index('Date')['Cases'].plot(title="Casos de Covid-19 en España desde 20/01/2020 hasta 06/07/2022")
Repetir el proceso para Panamá
Llevo a cabo el mismo código de variables, pero para la URL de Panamá.
df[df['Country'] == 'Panama']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 190 | Panama | panama | PA |
url_rt_pa = 'https://api.covid19api.com/country/panama/status/confirmed/live'
url_rt_pa
'https://api.covid19api.com/country/panama/status/confirmed/live'
df_rt_pa = pd.read_json(url_rt_pa)
df_rt_pa
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Panama | PA | 8.54 | -80.78 | 922990 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
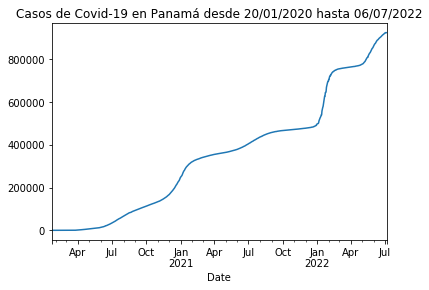
plot_rt_pa = df_rt_pa.set_index('Date')['Cases'].plot(title="Casos de Covid-19 en Panamá desde 20/01/2020 hasta 06/07/2022")
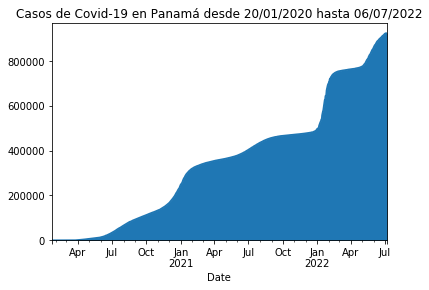
Añadiendo ,kind="area" al final se pondría el gráfico de área y con ,kind="bar" el gráfico de barras, aunque este formato no es lógico para este tipo de datos como puede visualizarse en el segundo ejemplo.
plot_rt_pa = df_rt_pa.set_index('Date')['Cases'].plot(title="Casos de Covid-19 en Panamá desde 20/01/2020 hasta 06/07/2022",kind="area")
plot_rt_pa = df_rt_pa.set_index('Date')['Cases'].plot(title="Casos de Covid-19 en Panamá desde 20/01/2020 hasta 06/07/2022",kind="bar")
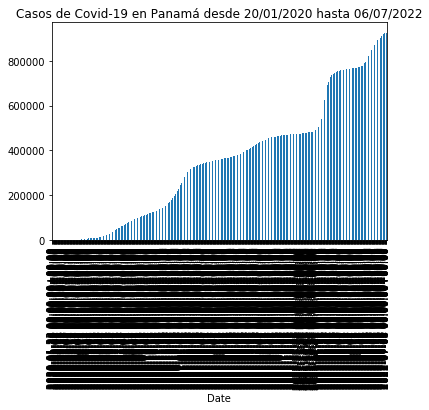
Ploteamos dos países
Para plotear dos o más paises hay que seguir los siguientes pasos:
Volvemos a identificar las variables previas de lectura de Json de las dos URLs para no tener que repetir el proceso desde arriba al cerrar el documento.
df_rt_pa = pd.read_json(url_rt_pa)
df_rt_es = pd.read_json(url_rt_es)
Se separan los casos por fecha en cada uno de los países y se plotea para visualizar la gráfica.
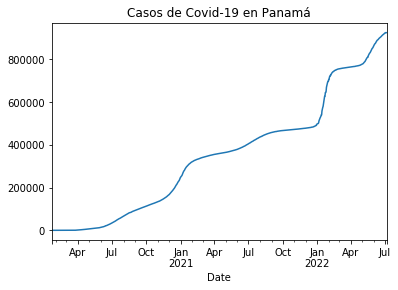
casos_pa = df_rt_pa.set_index('Date')['Cases']
casos_pa.plot(title="Casos de Covid-19 en Panamá")
<matplotlib.axes._subplots.AxesSubplot at 0x1f40fd89188>
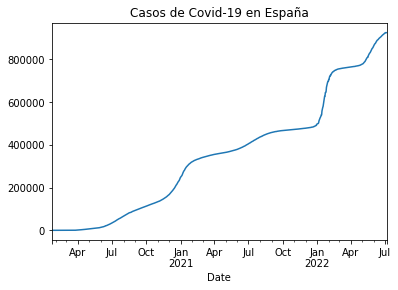
casos_es = df_rt_es.set_index('Date')['Cases']
casos_pa.plot(title="Casos de Covid-19 en España")
<matplotlib.axes._subplots.AxesSubplot at 0x1f41097d108>
Para visualizar podemos concatenar ambas tablas por fechas y casos en ambos países.
pa_vs_es = pd.concat([casos_es,casos_pa],axis=1)
pa_vs_es
| Cases | Cases | |
|---|---|---|
| Date | ||
| 2020-01-22 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-23 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-24 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-25 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-26 00:00:00+00:00 | 0.0 | 0 |
| ... | ... | ... |
| 2022-07-02 00:00:00+00:00 | 12818184.0 | 922990 |
| 2022-07-03 00:00:00+00:00 | 12818184.0 | 925254 |
| 2022-07-04 00:00:00+00:00 | 12818184.0 | 925254 |
| 2022-07-05 00:00:00+00:00 | 12890002.0 | 925254 |
| 2022-07-06 00:00:00+00:00 | NaN | 925254 |
897 rows × 2 columns
Para identificar los países marcamos las columnas con el nombre del país y visualizamos la gráfica con los datos nombrados.
pa_vs_es.columns= ['España','Panamá']
pa_vs_es
| España | Panamá | |
|---|---|---|
| Date | ||
| 2020-01-22 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-23 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-24 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-25 00:00:00+00:00 | 0.0 | 0 |
| 2020-01-26 00:00:00+00:00 | 0.0 | 0 |
| ... | ... | ... |
| 2022-07-02 00:00:00+00:00 | 12818184.0 | 922990 |
| 2022-07-03 00:00:00+00:00 | 12818184.0 | 925254 |
| 2022-07-04 00:00:00+00:00 | 12818184.0 | 925254 |
| 2022-07-05 00:00:00+00:00 | 12890002.0 | 925254 |
| 2022-07-06 00:00:00+00:00 | NaN | 925254 |
897 rows × 2 columns
Posteriormente a esta concatenación le hacemos un ploteo para visualizar la comparativa de forma visual.
pa_vs_es.plot(title="Comparativa Covid-19 España-Panamá")
<matplotlib.axes._subplots.AxesSubplot at 0x1f410997848>
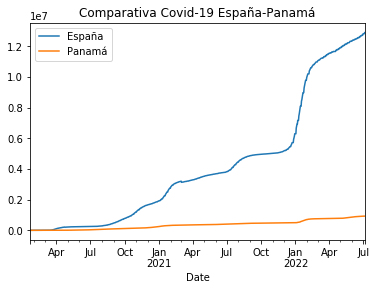
Si nos queremos descargar los datos en CSV podemos hacerlo con la función: pa_vs_es.to_csv('pa_vs_cr.csv') siendo pa_vs_cr el nombre de la tabla comparativa.
pa_vs_es.to_csv('pa_vs_cr.csv')
%ls
El volumen de la unidad C es Windows
El n£mero de serie del volumen es: 0699-4041
Directorio de C:\Users\RNASA\IriaSantos-web
06/07/2022 12:59 <DIR> .
06/07/2022 12:59 <DIR> ..
30/06/2022 19:23 <DIR> .ipynb_checkpoints
17/06/2022 22:31 6.774 ad1.md
22/06/2022 01:20 4.455 ad2.md
28/06/2022 01:08 93.323 ad3.ipynb
28/06/2022 01:11 75.389 ad3.md
27/06/2022 00:56 19.134 ad3_2.ipynb
27/06/2022 01:00 12.909 ad3_2.md
06/07/2022 12:59 477.300 api-covid-pandas.ipynb
06/07/2022 12:52 60.070 api-covid-pandas.md
06/07/2022 12:51 76.963 casos_pa_cr_ni_gt_sv_hn.svg
06/07/2022 12:52 13.901 output_22_0.png
06/07/2022 12:52 14.409 output_29_0.png
06/07/2022 12:52 13.425 output_31_0.png
06/07/2022 12:52 22.614 output_32_0.png
06/07/2022 12:52 12.117 output_38_1.png
06/07/2022 12:52 12.101 output_39_1.png
06/07/2022 12:52 14.743 output_45_1.png
06/07/2022 12:52 13.682 output_49_1.png
06/07/2022 12:52 13.682 output_50_1.png
06/07/2022 12:52 12.125 output_69_1.png
06/07/2022 12:52 12.303 output_70_1.png
06/07/2022 12:52 13.059 output_71_1.png
06/07/2022 12:52 12.429 output_72_1.png
06/07/2022 12:52 13.384 output_73_1.png
06/07/2022 12:52 12.255 output_74_1.png
06/07/2022 12:52 29.323 output_78_1.png
06/07/2022 12:52 27.315 output_81_1.png
06/07/2022 12:59 38.587 pa_vs_cr.csv
05/07/2022 20:28 15.518 pa_vs_es.png
05/07/2022 20:37 46.261 pa_vs_es.svg
30/06/2022 20:51 531 README.md
30 archivos 1.190.081 bytes
3 dirs 93.324.439.552 bytes libres
Para exportar una gráfica determinada en PNG o SVG seguiríamos los siguientes códigos:
import matplotlib.pyplot as plt
pa_vs_es.plot(title="Panamá vs España")
plt.savefig('pa_vs_es.png')
%ls
El volumen de la unidad C es Windows
El n£mero de serie del volumen es: 0699-4041
Directorio de C:\Users\RNASA\IriaSantos-web
06/07/2022 12:59 <DIR> .
06/07/2022 12:59 <DIR> ..
30/06/2022 19:23 <DIR> .ipynb_checkpoints
17/06/2022 22:31 6.774 ad1.md
22/06/2022 01:20 4.455 ad2.md
28/06/2022 01:08 93.323 ad3.ipynb
28/06/2022 01:11 75.389 ad3.md
27/06/2022 00:56 19.134 ad3_2.ipynb
27/06/2022 01:00 12.909 ad3_2.md
06/07/2022 12:59 477.300 api-covid-pandas.ipynb
06/07/2022 12:52 60.070 api-covid-pandas.md
06/07/2022 12:51 76.963 casos_pa_cr_ni_gt_sv_hn.svg
06/07/2022 12:52 13.901 output_22_0.png
06/07/2022 12:52 14.409 output_29_0.png
06/07/2022 12:52 13.425 output_31_0.png
06/07/2022 12:52 22.614 output_32_0.png
06/07/2022 12:52 12.117 output_38_1.png
06/07/2022 12:52 12.101 output_39_1.png
06/07/2022 12:52 14.743 output_45_1.png
06/07/2022 12:52 13.682 output_49_1.png
06/07/2022 12:52 13.682 output_50_1.png
06/07/2022 12:52 12.125 output_69_1.png
06/07/2022 12:52 12.303 output_70_1.png
06/07/2022 12:52 13.059 output_71_1.png
06/07/2022 12:52 12.429 output_72_1.png
06/07/2022 12:52 13.384 output_73_1.png
06/07/2022 12:52 12.255 output_74_1.png
06/07/2022 12:52 29.323 output_78_1.png
06/07/2022 12:52 27.315 output_81_1.png
06/07/2022 12:59 38.587 pa_vs_cr.csv
06/07/2022 12:59 15.524 pa_vs_es.png
05/07/2022 20:37 46.261 pa_vs_es.svg
30/06/2022 20:51 531 README.md
30 archivos 1.190.087 bytes
3 dirs 93.324.505.088 bytes libres
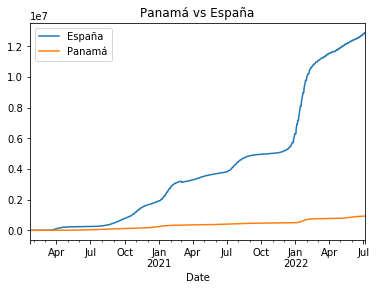
pa_vs_es.plot(title="Panamá vs España")
plt.savefig("pa_vs_es.svg", format="svg")
%ls
El volumen de la unidad C es Windows
El n£mero de serie del volumen es: 0699-4041
Directorio de C:\Users\RNASA\IriaSantos-web
06/07/2022 12:59 <DIR> .
06/07/2022 12:59 <DIR> ..
30/06/2022 19:23 <DIR> .ipynb_checkpoints
17/06/2022 22:31 6.774 ad1.md
22/06/2022 01:20 4.455 ad2.md
28/06/2022 01:08 93.323 ad3.ipynb
28/06/2022 01:11 75.389 ad3.md
27/06/2022 00:56 19.134 ad3_2.ipynb
27/06/2022 01:00 12.909 ad3_2.md
06/07/2022 12:59 477.300 api-covid-pandas.ipynb
06/07/2022 12:52 60.070 api-covid-pandas.md
06/07/2022 12:51 76.963 casos_pa_cr_ni_gt_sv_hn.svg
06/07/2022 12:52 13.901 output_22_0.png
06/07/2022 12:52 14.409 output_29_0.png
06/07/2022 12:52 13.425 output_31_0.png
06/07/2022 12:52 22.614 output_32_0.png
06/07/2022 12:52 12.117 output_38_1.png
06/07/2022 12:52 12.101 output_39_1.png
06/07/2022 12:52 14.743 output_45_1.png
06/07/2022 12:52 13.682 output_49_1.png
06/07/2022 12:52 13.682 output_50_1.png
06/07/2022 12:52 12.125 output_69_1.png
06/07/2022 12:52 12.303 output_70_1.png
06/07/2022 12:52 13.059 output_71_1.png
06/07/2022 12:52 12.429 output_72_1.png
06/07/2022 12:52 13.384 output_73_1.png
06/07/2022 12:52 12.255 output_74_1.png
06/07/2022 12:52 29.323 output_78_1.png
06/07/2022 12:52 27.315 output_81_1.png
06/07/2022 12:59 38.587 pa_vs_cr.csv
06/07/2022 12:59 15.524 pa_vs_es.png
06/07/2022 12:59 46.252 pa_vs_es.svg
30/06/2022 20:51 531 README.md
30 archivos 1.190.078 bytes
3 dirs 93.324.500.992 bytes libres
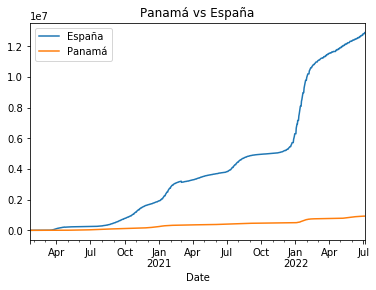
Con el programa de acceso libre Inkscape podemos realizar modificaciones en las gráficas en formato SVG (formato vectorial para la web).
Repetimos para lograr una comparativa de Centroamérica
El listado de países es:
- Panamá
- Costa Rica
- Nicaragua
- Guatemala
- El Salvador
- Honduras
Verifico cual es el slug y el ISO2 para emplear en la URL y en el identificador posteriormente (en todos los países).
df[df['Country'] == 'Panama']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 190 | Panama | panama | PA |
df[df['Country'] == 'Costa Rica']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 242 | Costa Rica | costa-rica | CR |
df[df['Country'] == 'Nicaragua']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 36 | Nicaragua | nicaragua | NI |
df[df['Country'] == 'Guatemala']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 239 | Guatemala | guatemala | GT |
df[df['Country'] == 'El Salvador']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 139 | El Salvador | el-salvador | SV |
df[df['Country'] == 'Honduras']
| Country | Slug | ISO2 | |
|---|---|---|---|
| 91 | Honduras | honduras | HN |
Defino la variable con la URL de cada país y hago una llamada a Json para esas URLs en busca de las tablas de datos.
url_rt_pa = 'https://api.covid19api.com/country/panama/status/confirmed/live'
df_rt_pa = pd.read_json(url_rt_pa)
df_rt_pa
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Panama | PA | 8.54 | -80.78 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Panama | PA | 8.54 | -80.78 | 922990 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Panama | PA | 8.54 | -80.78 | 925254 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
url_rt_cr = 'https://api.covid19api.com/country/costa-rica/status/confirmed/live'
df_rt_cr = pd.read_json(url_rt_cr)
df_rt_cr
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Costa Rica | CR | 9.75 | -83.75 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Costa Rica | CR | 9.75 | -83.75 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Costa Rica | CR | 9.75 | -83.75 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Costa Rica | CR | 9.75 | -83.75 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Costa Rica | CR | 9.75 | -83.75 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Costa Rica | CR | 9.75 | -83.75 | 904934 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Costa Rica | CR | 9.75 | -83.75 | 904934 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Costa Rica | CR | 9.75 | -83.75 | 904934 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Costa Rica | CR | 9.75 | -83.75 | 904934 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Costa Rica | CR | 9.75 | -83.75 | 904934 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
url_rt_ni = 'https://api.covid19api.com/country/nicaragua/status/confirmed/live'
df_rt_ni = pd.read_json(url_rt_ni)
df_rt_ni
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Nicaragua | NI | 12.87 | -85.21 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Nicaragua | NI | 12.87 | -85.21 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Nicaragua | NI | 12.87 | -85.21 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Nicaragua | NI | 12.87 | -85.21 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Nicaragua | NI | 12.87 | -85.21 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Nicaragua | NI | 12.87 | -85.21 | 14690 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Nicaragua | NI | 12.87 | -85.21 | 14690 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Nicaragua | NI | 12.87 | -85.21 | 14690 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Nicaragua | NI | 12.87 | -85.21 | 14690 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Nicaragua | NI | 12.87 | -85.21 | 14690 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
url_rt_gt = 'https://api.covid19api.com/country/guatemala/status/confirmed/live'
df_rt_gt = pd.read_json(url_rt_gt)
df_rt_gt
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Guatemala | GT | 15.78 | -90.23 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Guatemala | GT | 15.78 | -90.23 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Guatemala | GT | 15.78 | -90.23 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Guatemala | GT | 15.78 | -90.23 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Guatemala | GT | 15.78 | -90.23 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Guatemala | GT | 15.78 | -90.23 | 918797 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Guatemala | GT | 15.78 | -90.23 | 920294 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Guatemala | GT | 15.78 | -90.23 | 921146 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Guatemala | GT | 15.78 | -90.23 | 922340 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Guatemala | GT | 15.78 | -90.23 | 922340 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
url_rt_sv = 'https://api.covid19api.com/country/el-salvador/status/confirmed/live'
df_rt_sv = pd.read_json(url_rt_sv)
df_rt_sv
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | El Salvador | SV | 13.79 | -88.9 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | El Salvador | SV | 13.79 | -88.9 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | El Salvador | SV | 13.79 | -88.9 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | El Salvador | SV | 13.79 | -88.9 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | El Salvador | SV | 13.79 | -88.9 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | El Salvador | SV | 13.79 | -88.9 | 169646 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | El Salvador | SV | 13.79 | -88.9 | 169646 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | El Salvador | SV | 13.79 | -88.9 | 169646 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | El Salvador | SV | 13.79 | -88.9 | 169646 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | El Salvador | SV | 13.79 | -88.9 | 169646 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
url_rt_hn = 'https://api.covid19api.com/country/honduras/status/confirmed/live'
df_rt_hn = pd.read_json(url_rt_hn)
df_rt_hn
| Country | CountryCode | Province | City | CityCode | Lat | Lon | Cases | Status | Date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Honduras | HN | 15.2 | -86.24 | 0 | confirmed | 2020-01-22 00:00:00+00:00 | |||
| 1 | Honduras | HN | 15.2 | -86.24 | 0 | confirmed | 2020-01-23 00:00:00+00:00 | |||
| 2 | Honduras | HN | 15.2 | -86.24 | 0 | confirmed | 2020-01-24 00:00:00+00:00 | |||
| 3 | Honduras | HN | 15.2 | -86.24 | 0 | confirmed | 2020-01-25 00:00:00+00:00 | |||
| 4 | Honduras | HN | 15.2 | -86.24 | 0 | confirmed | 2020-01-26 00:00:00+00:00 | |||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 892 | Honduras | HN | 15.2 | -86.24 | 427718 | confirmed | 2022-07-02 00:00:00+00:00 | |||
| 893 | Honduras | HN | 15.2 | -86.24 | 427718 | confirmed | 2022-07-03 00:00:00+00:00 | |||
| 894 | Honduras | HN | 15.2 | -86.24 | 427718 | confirmed | 2022-07-04 00:00:00+00:00 | |||
| 895 | Honduras | HN | 15.2 | -86.24 | 427718 | confirmed | 2022-07-05 00:00:00+00:00 | |||
| 896 | Honduras | HN | 15.2 | -86.24 | 427718 | confirmed | 2022-07-06 00:00:00+00:00 |
897 rows × 10 columns
Selecciono para cada una de las tablas los casos por fecha y ploteo la gráfica.
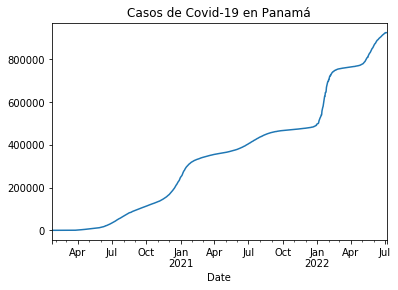
casos_pa = df_rt_pa.set_index('Date')['Cases']
casos_pa.plot(title="Casos de Covid-19 en Panamá")
<matplotlib.axes._subplots.AxesSubplot at 0x1f4105ea448>
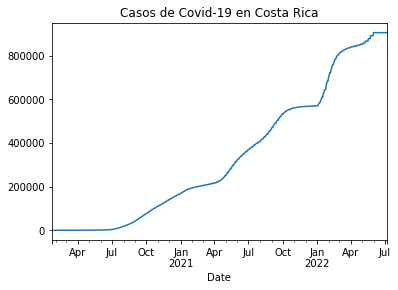
casos_cr = df_rt_cr.set_index('Date')['Cases']
casos_cr.plot(title="Casos de Covid-19 en Costa Rica")
<matplotlib.axes._subplots.AxesSubplot at 0x1f411c76108>
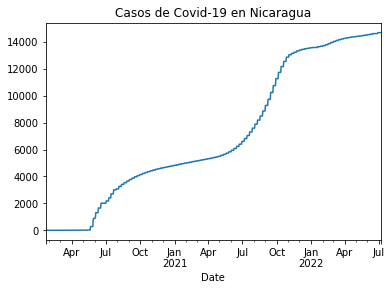
casos_ni = df_rt_ni.set_index('Date')['Cases']
casos_ni.plot(title="Casos de Covid-19 en Nicaragua")
<matplotlib.axes._subplots.AxesSubplot at 0x1f411cfff88>
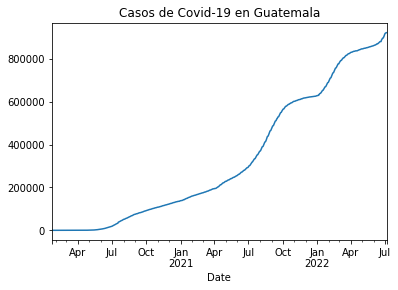
casos_gt = df_rt_gt.set_index('Date')['Cases']
casos_gt.plot(title="Casos de Covid-19 en Guatemala")
<matplotlib.axes._subplots.AxesSubplot at 0x1f411d69088>
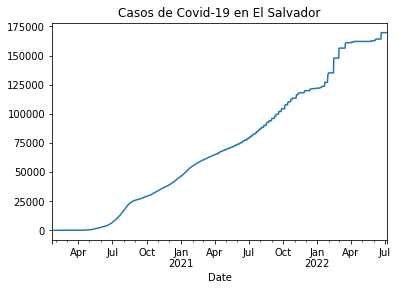
casos_sv = df_rt_sv.set_index('Date')['Cases']
casos_sv.plot(title="Casos de Covid-19 en El Salvador")
<matplotlib.axes._subplots.AxesSubplot at 0x1f411c28bc8>
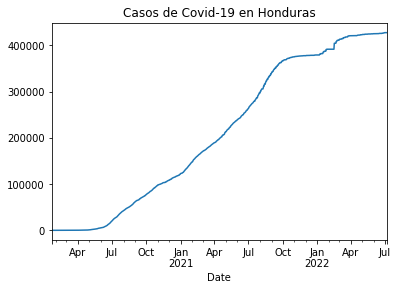
casos_hn = df_rt_hn.set_index('Date')['Cases']
casos_hn.plot(title="Casos de Covid-19 en Honduras")
<matplotlib.axes._subplots.AxesSubplot at 0x1f412060848>
Concateno todas las tablas previas y las nombro por cada uno de los países a los que pertenecen las columnas.
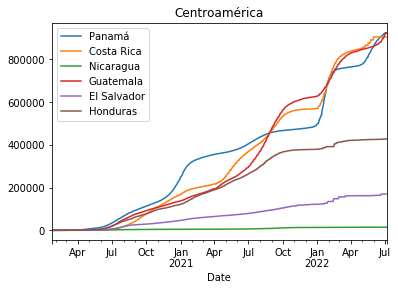
casos_pa_cr_ni_gt_sv_hn = pd.concat([casos_pa,casos_cr,casos_ni,casos_gt,casos_sv,casos_hn],axis=1)
casos_pa_cr_ni_gt_sv_hn.columns= ['Panamá','Costa Rica', 'Nicaragua', 'Guatemala', 'El Salvador', 'Honduras']
casos_pa_cr_ni_gt_sv_hn
| Panamá | Costa Rica | Nicaragua | Guatemala | El Salvador | Honduras | |
|---|---|---|---|---|---|---|
| Date | ||||||
| 2020-01-22 00:00:00+00:00 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-23 00:00:00+00:00 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-24 00:00:00+00:00 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-25 00:00:00+00:00 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-26 00:00:00+00:00 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... |
| 2022-07-02 00:00:00+00:00 | 922990 | 904934 | 14690 | 918797 | 169646 | 427718 |
| 2022-07-03 00:00:00+00:00 | 925254 | 904934 | 14690 | 920294 | 169646 | 427718 |
| 2022-07-04 00:00:00+00:00 | 925254 | 904934 | 14690 | 921146 | 169646 | 427718 |
| 2022-07-05 00:00:00+00:00 | 925254 | 904934 | 14690 | 922340 | 169646 | 427718 |
| 2022-07-06 00:00:00+00:00 | 925254 | 904934 | 14690 | 922340 | 169646 | 427718 |
897 rows × 6 columns
Posteriormente ploteo para visualizar la gráfica comparativa.
casos_pa_cr_ni_gt_sv_hn.plot(title="Comparativa Covid-19 en los países de Centroamérica")
<matplotlib.axes._subplots.AxesSubplot at 0x1f4105fb948>
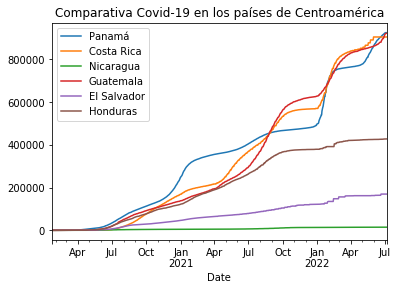
Para finalizar, exporto la tabla en CSV y la gráfica en formato vectorial para web (SVG).
casos_pa_cr_ni_gt_sv_hn.to_csv('casos_pa_cr_ni_gt_sv_hn.csv')
%ls
El volumen de la unidad C es Windows
El n£mero de serie del volumen es: 0699-4041
Directorio de C:\Users\RNASA\IriaSantos-web
06/07/2022 13:00 <DIR> .
06/07/2022 13:00 <DIR> ..
30/06/2022 19:23 <DIR> .ipynb_checkpoints
17/06/2022 22:31 6.774 ad1.md
22/06/2022 01:20 4.455 ad2.md
28/06/2022 01:08 93.323 ad3.ipynb
28/06/2022 01:11 75.389 ad3.md
27/06/2022 00:56 19.134 ad3_2.ipynb
27/06/2022 01:00 12.909 ad3_2.md
06/07/2022 12:59 477.300 api-covid-pandas.ipynb
06/07/2022 12:52 60.070 api-covid-pandas.md
06/07/2022 13:00 56.543 casos_pa_cr_ni_gt_sv_hn.csv
06/07/2022 12:51 76.963 casos_pa_cr_ni_gt_sv_hn.svg
06/07/2022 12:52 13.901 output_22_0.png
06/07/2022 12:52 14.409 output_29_0.png
06/07/2022 12:52 13.425 output_31_0.png
06/07/2022 12:52 22.614 output_32_0.png
06/07/2022 12:52 12.117 output_38_1.png
06/07/2022 12:52 12.101 output_39_1.png
06/07/2022 12:52 14.743 output_45_1.png
06/07/2022 12:52 13.682 output_49_1.png
06/07/2022 12:52 13.682 output_50_1.png
06/07/2022 12:52 12.125 output_69_1.png
06/07/2022 12:52 12.303 output_70_1.png
06/07/2022 12:52 13.059 output_71_1.png
06/07/2022 12:52 12.429 output_72_1.png
06/07/2022 12:52 13.384 output_73_1.png
06/07/2022 12:52 12.255 output_74_1.png
06/07/2022 12:52 29.323 output_78_1.png
06/07/2022 12:52 27.315 output_81_1.png
06/07/2022 12:59 38.587 pa_vs_cr.csv
06/07/2022 12:59 15.524 pa_vs_es.png
06/07/2022 12:59 46.252 pa_vs_es.svg
30/06/2022 20:51 531 README.md
31 archivos 1.246.621 bytes
3 dirs 93.324.234.752 bytes libres
casos_pa_cr_ni_gt_sv_hn.plot(title="Centroamérica")
plt.savefig("casos_pa_cr_ni_gt_sv_hn.svg", format="svg")
%ls
El volumen de la unidad C es Windows
El n£mero de serie del volumen es: 0699-4041
Directorio de C:\Users\RNASA\IriaSantos-web
06/07/2022 13:00 <DIR> .
06/07/2022 13:00 <DIR> ..
30/06/2022 19:23 <DIR> .ipynb_checkpoints
17/06/2022 22:31 6.774 ad1.md
22/06/2022 01:20 4.455 ad2.md
28/06/2022 01:08 93.323 ad3.ipynb
28/06/2022 01:11 75.389 ad3.md
27/06/2022 00:56 19.134 ad3_2.ipynb
27/06/2022 01:00 12.909 ad3_2.md
06/07/2022 12:59 477.300 api-covid-pandas.ipynb
06/07/2022 12:52 60.070 api-covid-pandas.md
06/07/2022 13:00 56.543 casos_pa_cr_ni_gt_sv_hn.csv
06/07/2022 13:00 76.963 casos_pa_cr_ni_gt_sv_hn.svg
06/07/2022 12:52 13.901 output_22_0.png
06/07/2022 12:52 14.409 output_29_0.png
06/07/2022 12:52 13.425 output_31_0.png
06/07/2022 12:52 22.614 output_32_0.png
06/07/2022 12:52 12.117 output_38_1.png
06/07/2022 12:52 12.101 output_39_1.png
06/07/2022 12:52 14.743 output_45_1.png
06/07/2022 12:52 13.682 output_49_1.png
06/07/2022 12:52 13.682 output_50_1.png
06/07/2022 12:52 12.125 output_69_1.png
06/07/2022 12:52 12.303 output_70_1.png
06/07/2022 12:52 13.059 output_71_1.png
06/07/2022 12:52 12.429 output_72_1.png
06/07/2022 12:52 13.384 output_73_1.png
06/07/2022 12:52 12.255 output_74_1.png
06/07/2022 12:52 29.323 output_78_1.png
06/07/2022 12:52 27.315 output_81_1.png
06/07/2022 12:59 38.587 pa_vs_cr.csv
06/07/2022 12:59 15.524 pa_vs_es.png
06/07/2022 12:59 46.252 pa_vs_es.svg
30/06/2022 20:51 531 README.md
31 archivos 1.246.621 bytes
3 dirs 93.324.300.288 bytes libres